Identification of linezolid-resistant Enterococcus spp. in calves: findings from Portuguese high-yielding dairy farms
DOI:
https://doi.org/10.48797/sl.2025.324Keywords:
Selected Oral CommunicationAbstract
Background: Antimicrobial resistance (AMR) is a global health threat requiring a One Health approach, as antibiotic-resistant bacteria can spread between animals and humans. Enterococcus spp., particularly E. faecium (Efm) and E. faecalis (Efs), are key AMR indicators due to their role as gut commensals and their potential as reservoirs of resistance genes. They are also opportunistic pathogens that can cause severe human infections. Cattle are a recognized reservoir of multidrug-resistant (MDR) Enterococcus spp., yet remain among the least studied food-producing animals in this context [1,2]. Objective: To assess if contemporary faecal samples from main cattle farms in Northern Portugal carry clinically relevant antibiotic-resistant Enterococcus spp. Methods: Thirty bovine fecal swab samples were collected from 10 high-yielding farms with Holstein-Friesian dairy cattle in 2 cities during 2023 [3]. Sample processing included pre-enrichment (37ºC/18h) without/with antibiotics (ampicillin 16μg/mL, vancomycin 8μg/mL or florfenicol 16μg/mL) followed by plating onto Slanetz–Bartley selective agar, without/with the same antibiotics (37°C/48h). Typical colonies were saved for identification (MALDI-TOF MS) and antibiotic susceptibility test (disk diffusion; EUCAST/CLSI). Prevalence percentages were calculated on a per-sample basis. Results: All samples contained Enterococcus (n=43) that were identified as Efm (n=18, 60%), E. hirae (n=14, 47%), Efs (n=9, 30%), E. casseliflavus and E. durans (n=1 each, 3%); and resistant to erythromycin (44%), tetracycline (39%), chloramphenicol (20%), ampicillin, linezolid, high-level streptomycin (17% each), ciprofloxacin (13%), and high-level gentamicin (7%). MDR isolates (23%) were mostly obtained from calves rather than adults, and only found in Efs (67%) and Efm(50%) species. Linezolid-resistant isolates were only recovered from supplemented media with florfenicol while those resistant to ampicillin were better detected using culture medium with ampicillin or florfenicol. Ampicillin resistance was only detected in Efm while linezolid resistance was identified in both Efm and Efs (all calves, all MDR, 2 farms). Conclusions: Our study shows that dairy cattle carry MDR Enterococcus spp., including strains resistant to critically important antibiotics in the treatment of human infections (linezolid). These findings underscore the urgent need for sustained AMR surveillance and cross-sector collaboration within a One Health framework.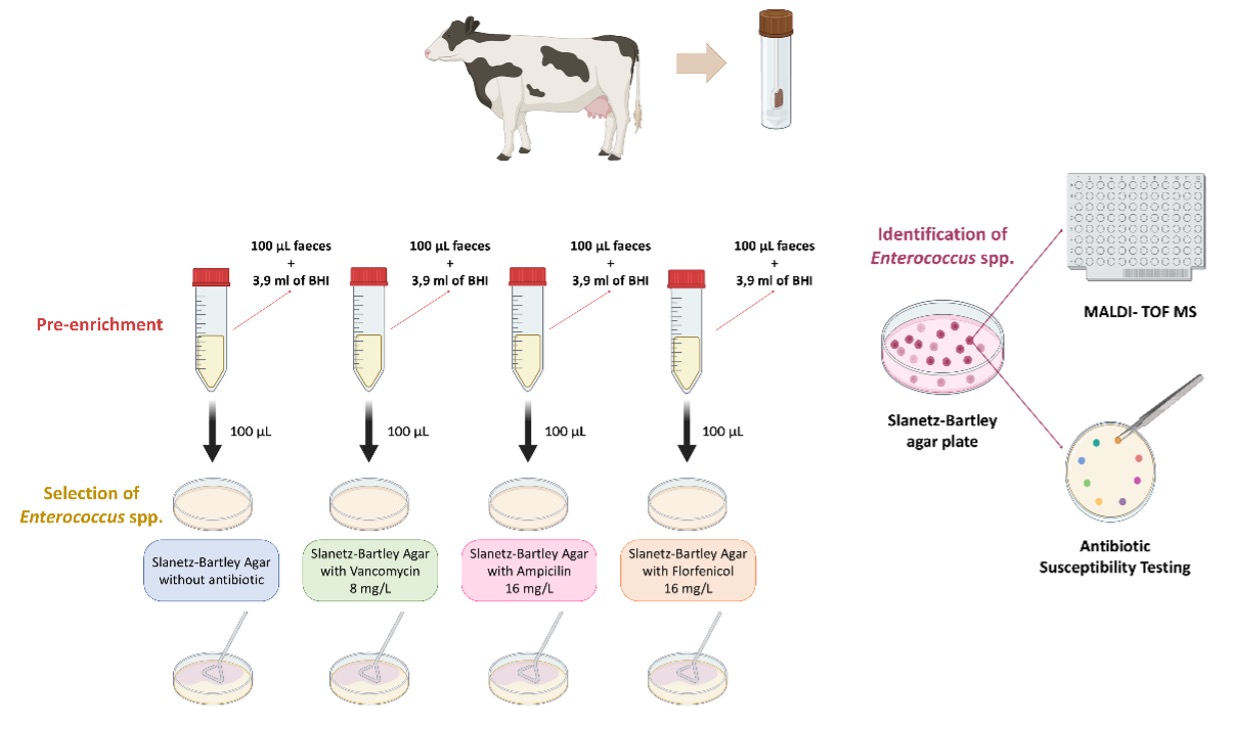
Figure 1. Schematic processing of cattle faecal swab samples towards the isolation, identification and characterization of Enterococcus spp.
References
1.The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2022–2023. EFSA Journal. 2025, 23(3), doi: 10.2903/j.efsa.2025.9237
2. Gião J, Leão C, Albuquerque T, Clemente L, Amaro A. Antimicrobial Susceptibility of Enterococcus Isolates from Cattle and Pigs in Portugal: Linezolid Resistance Genes optrA and poxtA. Antibiotics. 2022, 11(5), 615, doi: 10.3390/antibiotics11050615
3. Quinteira S, Dantas R, Pinho L, et al. Dairy Cattle and the Iconic Autochthonous Cattle in Northern Portugal Are Reservoirs of Multidrug-Resistant Escherichia coli. Antibiotics. 2024, 13(12), 1208, doi: 10.3390/antibiotics13121208
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Alícia Ribeiro, Maria J. Teixeira, Luís Pinho, Carla Campos, Carla Novais, Luísa Peixe, Nuno V. Brito, Sandra Quinteira, Carla Miranda, Ana R. Freitas

This work is licensed under a Creative Commons Attribution 4.0 International License.
In Scientific Letters, articles are published under a CC-BY license (Creative Commons Attribution 4.0 International License), the most open license available. The users can share (copy and redistribute the material in any medium or format) and adapt (remix, transform, and build upon the material for any purpose, even commercially), as long as they give appropriate credit, provide a link to the license, and indicate if changes were made (read the full text of the license terms and conditions of use).
The author is the owner of the copyright.









